Clear filter
Announcement
Anastasia Dyubaylo · Oct 19, 2021
Hey Developers,
Planning to attend the Focus Sessions of InterSystems Virtual Summit 2021? Don't miss the session dedicated to InterSystems Developer Community, Open Exchange & Global Masters!
⚡️ "Win. Win. Win with InterSystems Developer Ecosystem" VSummit21 session ⚡️
🎁 Note: All session attendees get a special prize.
Speakers: 🗣 @Anastasia.Dyubaylo, Community Manager, InterSystems 🗣 @Lena.Evsikova, Product Owner of InterSystems Open Exchange🗣 @Olga.Zavrazhnova2637, Customer Advocacy Manager, InterSystems
Learn how to succeed with the InterSystems Developer Community, Global Masters gamification hub, and Open Exchange application gallery.
Interests: Developer Experience, InterSystems IRIS, User Communities
Our Focus Sessions are available on-demand for viewing from Wednesday, October 27!
So!
Join our session to enjoy the full experience of InterSystems Technology and our Ecosystem for Developers! Looking forward to it!! I will be there! @Olga.Zavrazhnova2637@Lena.Evsikova @Anastasia.Dyubaylo Just watched your presentation. It was really GREAT !!You raised the level of presentation significantly.The most attractive presentation for me so far.It will be hard to top you.CONGRATULATIONS ! So happy to hear that, Robert! Thank you! I completely agree with @Robert.Cemper1003! Great job highlighting the D.C., OEX and Global Masters!! Thank you, Robert! So happy to hear such feedback 😊 Thank you, Ben ☺️ Very good presentation. They are a true trio of pocker aces to win
And all the crew that are in the same ship
Announcement
Anastasia Dyubaylo · Dec 15, 2021
Hey Developers,
Learn about the InterSystems Partner Directory, its value to partners, and how to join:
⏯ Partner Directory New Services for InterSystems Partners & End Users
🗣 Presenter: Elizabeth Zaylor, Partnerships & Alliances, InterSystems
See you at https://partner.intersystems.com!
Announcement
Olga Zavrazhnova · Dec 5, 2021
Hey Community,
This is the time to show our passion for the InterSystems Developer Community! We're so proud to announce that InterSystems Global Masters is a finalist for an Influitive BAMMIE Award for Most Passionate Community🤩🤩🤩
Certainly, this is because of you, our great community members!
BUT, for us to win, we'll need to get more votes than other finalists - so we need your votes!
🚀 PLEASE press "You've got my vote" > in this challenge <
Vote every day till December 9 to show that we have the most engaged community!
Let's WIN together!
3-days left! Video from Olga :)
Hey Community, only 3 days are left to vote for Global Masters! Community!
We really need your support!!! Please vote for us 🙏🏼 Vote Community! Vote, please! We have to support our dev community! Today is the last voting day! Please press the button for us in this challenge https://globalmasters.intersystems.com/challenges/2982Thank you, Community, for support! Fingers crossed!
Article
Botai Zhang · Jan 25, 2021
Built in multi model integration using InterSystems iris data platform
Solution of hospital information inquiry business
Integration of hospital information query business solutions using InterSystems IRIS data platform with built-in multiple models
### Summary:
With the gradual improvement of hospital information construction, there are more and more hospital subsystems, and more and more interfaces between systems. At the same time, the interface cost is increasing, and the management work is becoming more and more complex. Among them, the number of query business interfaces is gradually increasing according to the business type differentiation, which brings problems such as large amount of interfaces, heavy development work, code redundancy, difficult maintenance and so on. In view of this dilemma, we use InterSystems IRIS data platform built-in multi model integration of hospital information query business solutions. The application can be configured to complete the query business interface implementation, greatly reducing the key operation cycle of development, maintenance, implementation and other projects.Key applications: IRIS, Rest API, ObjectScript, Globals, SQL, data lookup tables
### Application model and Application Introduction:
1. Using the model 1. Globals (key-value) Globals is a sparse multidimensional array that can be stored and managed in IRIS database. You can use ObjectScript and native APIs to work with globals. **Tools:** https://docs.intersystems.com/irislatest/csp/docbook/DocBook.UI.Page.cls?KEY=GGBL_MANAGING **Application:** According to the key value pair of globals, the application program has the characteristics of fast access speed. It is applied in the rest dispatch class and BP process management of this program, which solves the problem of frequent value taking, slow speed and configuration operation on the front page of the lookup table, such as storing SQL model, service configuration information and so on. 2. SQL access InterSystems iris provides SQL access to data through ObjectScript, rest API and JDBC **Tools:** https://docs.intersystems.com/irislatest/csp/docbook/Doc.View.cls?KEY=GSQL_smp **Application:** In the query business, the three-party system does not cooperate with the interface transformation, which leads to the difficulty of interface implementation. At this time, we use iris embedded model ObjectScript, rest API and JDBC to realize SQL access to data and establish business interface. 3. Object access Through ObjectScript and rest API, InterSystems iris provides a way to store and change object instances in globals. **File:** https://docs.intersystems.com/irislatest/csp/docbook/Doc.View.cls?KEY=PAGE_multimodel_object **Application:** During the whole interaction process, the InterSystems iris object is manipulated directly. ObjectScript class definitions are often used as templates for creating objects such as patients, departments, or healthcare workers. 2. Establish application cases (this application takes patients as an example) 1. application construction: 1. basic environment Iris version information: iris for windows (x86-64) 2020.1 (build 215u) mon Mar 30 2020 20:14:33 EDT \[Health Connect:2.1.0] Iris has java and JDBC environment Postman can be used for testing 2. installation steps ① Establish rest Service New web application → configure dispatch class → configure permission. This step can be seen in the following pictures: application running / webreplication (query). PNG and webreplication (role). PNG ② Configure sql-jdbc Establish SQL connection, connect to test database mysql, import test jhip_ patient_ info.sql ③ Configuration lookup table Global-^ Ens.LookupTable Look up table file import in ④ Import code Import the code in applicationcode, compile and open production, Note: modify Bo configuration information (DNS), configure Java gateway, etc 2. application process Omitted (see PDF) 3. Application Test The postman tool (or other tools) can be used for test verification Postman can import Query.postman_ collection.json , change IP, port number information and URL for testing. 4. application summary This application takes patient service query as a case, which can be configured with inbound and outbound protocols, query conditions and business types to solve the problem of query business interface.
Finally, if you agree with this solution, please vote for the application in the inter systems multi model database contest.
Voting links
Application name:HealthInfoQueryLayer
Thank you very much for your support!
Announcement
Anastasia Dyubaylo · Mar 4, 2021
Hey Developers,
See how the InterSystems IRIS FHIR Server allows you to develop and deploy your FHIR applications on AWS without manual configuration and deployment:
⏯ Getting Started with the InterSystems IRIS FHIR Server on AWS
👉🏼 Subscribe to InterSystems Developers YouTube.
Enjoy and stay tuned!
Article
Yuri Marx · Dec 21, 2020
Today, is important analyze the content into portals and websites to get informed, analyze the concorrents, analyze trends, the richness and scope of content of websites. To do this, you can alocate people to read thousand of pages and spend much money or use a crawler to extract website content and execute NLP on it. You will get all necessary insights to analyze and make precise decisions in a few minutes.
Gartner defines web crawler as: "A piece of software (also called a spider) designed to follow hyperlinks to their completion and to return to previously visited Internet addresses".
There are many web crawlers to extract all relevant website content. In this article I present to you Crawler4J. It is the most used software to extract website content and has MIT license. Crawler4J needs only the root URL, the depth (how many child sites will be visited) and total pages (if you want limit the pages extracted). By default only textual content will be extracted, but you config the engine to extract all website files!
I created a PEX Java service to allows you using an IRIS production to extract the textual content to any website. the content is stored into a local folder and the IRIS NLP reads these files and show to you all text analytics insights!
To see it in action follow these procedures:
1 - Go to https://openexchange.intersystems.com/package/website-analyzer and click Download button to see app github repository.
2 - Create a local folder in your machine and execute: https://github.com/yurimarx/website-analyzer.git.
3 - Go to the project directory: cd website-analyzer.
4 - Execute: docker-compose build (wait some minutes)
5 - Execute: docker-compose up -d
6 - Open your local InterSystems IRIS: http://localhost:52773/csp/sys/UtilHome.csp (user _SYSTEM and password SYS)
7 - Open the production and start it: http://localhost:52773/csp/irisapp/EnsPortal.ProductionConfig.zen?PRODUCTION=dc.WebsiteAnalyzer.WebsiteAnalyzerProduction
8 - Now, go to your browser to initiate a crawler: http://localhost:9980?Website=https://www.intersystems.com/ (to analyze intersystems site, any URL can be used)
9 - Wait between 40 and 60 seconds. A message you be returned (extracted with success). See above sample.
10 - Now go to Text Analytics to analyze the content extracted: http://localhost:52773/csp/IRISAPP/_iKnow.UI.KnowledgePortal.zen?$NAMESPACE=IRISAPP&domain=1
11 - Return to the production and see Depth and TotalPages parameters, increase the values if you want extract more content. Change Depth to analyze sub links and change TotalPages to analyze more pages.
12 - Enjoy! And if you liked, vote (https://openexchange.intersystems.com/contest/current) in my app: website-analyzer
I will write a part 2 with implementations details, but all source code is available in Github. Hi Yuri!Very interesting app!But as I am not a developer, could you please tell more about the results the analizer will give to a marketer or a website owner? Which insights could be extracted form the analysis? Hi @Elena.E
I published a new article about marketing and this app: https://community.intersystems.com/post/marketing-analysis-intersystems-website-using-website-analyzer
About the possible results allows you:
1. Get the most popular words, terms and sentences wrote into the website, so you discover the business focus, editorial line and marketing topics.
2. Sentiment analysis into the sentences, the content is has positive or negative focus
3. Rich cloud words to all the website. Rich because is a semantic analysis, with links between words and sentences
4. Dominance and frequence analysis, to analyze trends
5. Connections paths between sentences, to analyze depth and coverage about editorial topics
6. Search engine of topics covered, the website discuss a topic? How many times do this?
7. Product analysis, the app segment product names and link the all other analysis, so you can know if the website says about your product and Services and the frequency Hi Yuri!
This is a fantastic app!
And works!
But the way to set up the crawler is not that convenient and not very user-friendly.
You never know if the crawler works and if you placed the URL right.
Is it possible to add a page which will let you place the URL, start/stop crawler and display some progress if any?
Maybe I ask a lot :)
Anyway, this is a really great tool to perform IRIS NLP vs ANY site:
Announcement
Anastasia Dyubaylo · Dec 12, 2020
Hey Developers,
Please welcome the new video by @sween on InterSystems Developers YouTube:
⏯ InterSystems IRIS Native Python API in AWS Lambda
In this video, you will learn the seamless InterSystems IRIS functionality in the AWS Serverless ecosystem.
Leave your questions in the comments to this post.
Enjoy and stay tuned!
Announcement
Olga Zavrazhnova · Dec 14, 2020
Hi Developers,
We invite you to take a few minutes and leave a review about your experience with InterSystems IRIS on the TrustRadius. Submitting a review requires time and effort, so we'll be glad to reward you with a $25 VISA Gift Card for a published review! UPDATE: this promotion ended in 2021.
To get a $25 VISA card from InterSystems follow these steps:
✅ #1: Follow → this link ← to submit a review (click on the "Start My Review" button).
✅ #2: Your review will have a title (headline). Copy the text of the headline of your review and paste it in this challenge on Global Masters.
✅ #3: After your review is published you will get the $25 VISA Cards and 3000 points on Global Masters.
Please note:
TrustRadius must approve the survey to qualify for the gift card. TrustRadius will not approve reviews from resellers, systems integrators, or MSP/ISV’s of InterSystems.
TrustRadius does not approve the reviews that have been previously published online.
Done? Awesome! Your gift card is on the way!
Hmm. The link to GM says:
Ooops!
Sorry friend, looks like this challenge is no longer available.
My title: " Never say IMPOSSIBLE with IRIS "
Hi Robert, thank you for submitting a review for us! I made some corrections to the challenge, so the link should work for you now. THX. Just verified it. Hi, the GM link is broken. It says:
Ooops!
Sorry friend, looks like this challenge is no longer available. here it is:https://www.trustradius.com/reviews/intersystems-iris-2020-12-15-16-53-48 Hi Akshay,I see you registered recently - welcome to Global Masters! Could you please complete this challenge first? After it's done the TrustRadius challenge will be unlocked for you. Thank you, Robert! Your gift card is already opened for you on GM Thanks for your help. Here's the published review! https://www.trustradius.com/reviews/intersystems-iris-2020-12-23-14-00-04 Hi Akshay! So great, thank you! Your reward is opened for you on Global Masters. Happy New Year! There seems to be some error. I clicked on Redeem, and my card just disappeared. It is not showing in the Rewards section either! That's the way it works. You consume it once.You'll get a mail once processed. But probably not this year Ah no issues, I'll wait until next year. I've got nothing but time. Thanks though!! I can't see any challenges... What do I do now? Great Unable to paste in challenge on Global Master
Article
Mihoko Iijima · Mar 5, 2021
**This article is a continuation of this post.**
In the previous article, we discussed the development of business processes, which are part of the components required for system integration and serve as a production coordinator.
This article will discuss creating a business service, which is the information input window for production.
* Production
* Message
* **Components**
* **Business services**
* Business processes(previous post)
* Business operation
And finally, the last component of “Let's Use Interoperability!”
The business service provides a window of input for information sent from outside IRIS, with or without using the adapter for external I/F.
There are three types of business services in the sample (links in parentheses are links to the sample code):
1. [Business services for files using inbound adapters](#fileinboundadapter)(Start.FileBS)
2. [Business services for Web services using the SOAP inbound adapter](#soapinboundadapter)(Start.WS.WebServiceBS)
3. [Business service called by stored procedure or REST without using an adapter](#nonadapter)(Start.NonAdapterBS)
Different connection methods used for inputting information will only increase the number of business services; however, the processing done within a business service is
Create a request message to be sentusing externally inputted information and simply call the business component
It's effortless.
Now, let's outline how to create components that use file-inbound adapters.
Business services are written in scripts, which can be created in VSCode or Studio (see this article on using VSCode).
### 1. Business services for files using inbound adapters(Start.FileBS)
If you create a class in VSCode, you should create a class that inherits from Ens.BusinessService. As for adapters, you can use the **ADAPTER** parameter as ADAPTER class name (e.g., specify a file-inbound adapter class).
If you do not use the adapter, no configuration is required.
```objectscript
Class Start.FileBS Extends Ens.BusinessService
{
Parameter ADAPTER = "EnsLib.File.InboundAdapter";
```
In the file-inbound adapter, you can specify the directory to be monitored in Settings→File Path for the production's business service.
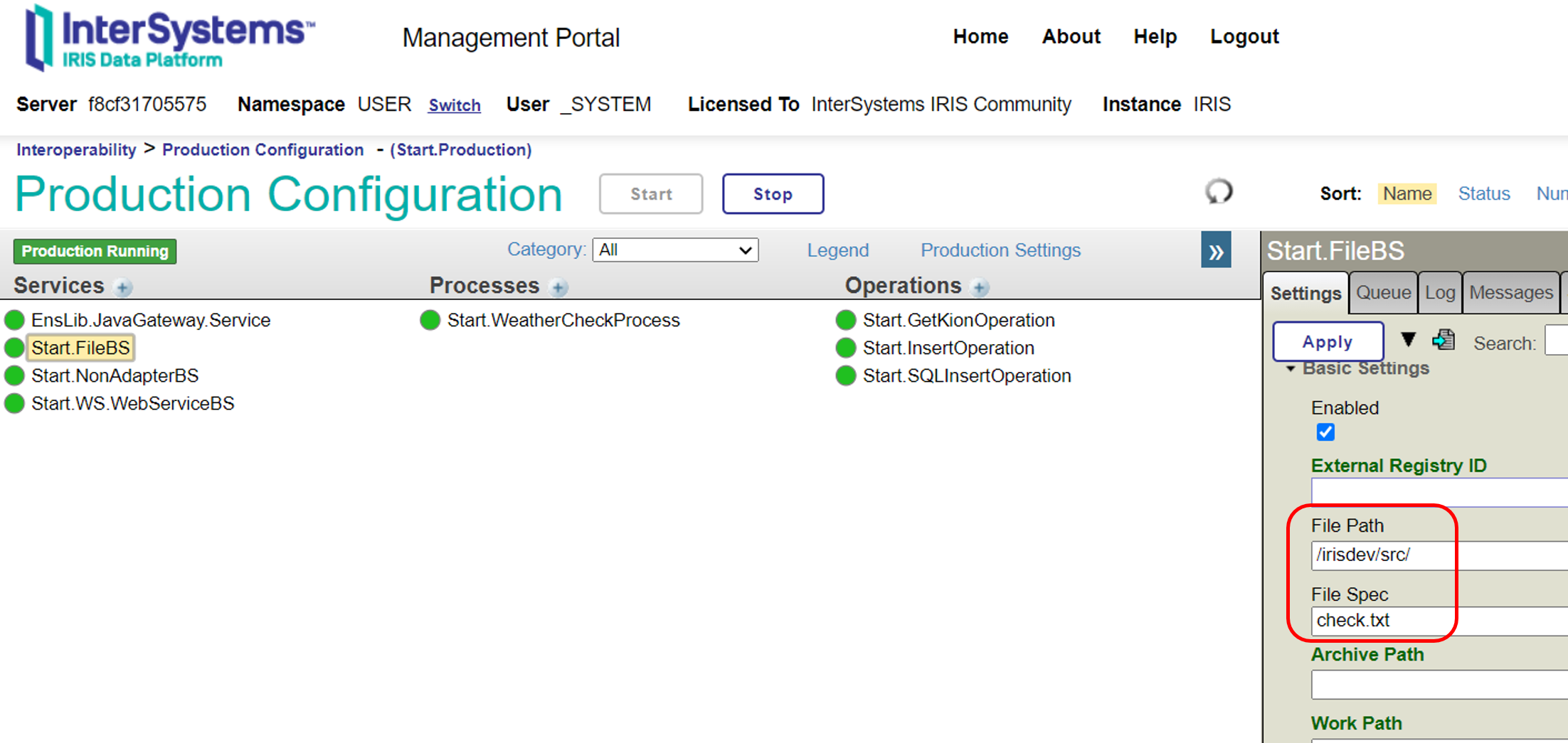
If the file located in the "File Path" matches the information specified in the "File Spec," it opens the file as a stream object. It defines it as the first variable when calling the business service **ProcessInput()**.
When **ProcessInput()** is started, **OnProcessInput()** is automatically called. OnProcessInput() is passed directly to **OnProcessInput()** with the parameters passed to ProcessInput().
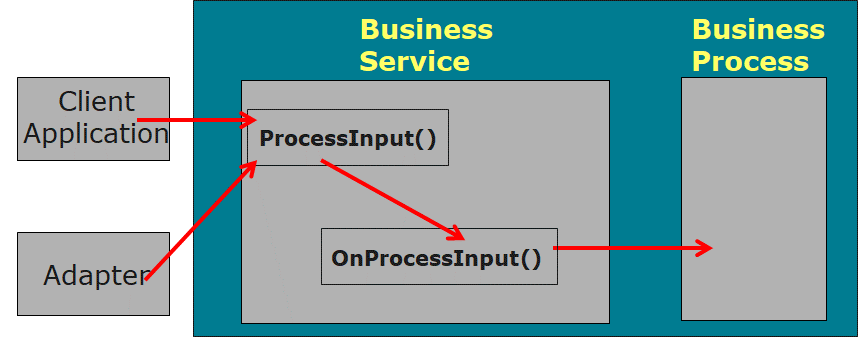
In **OnProcessInput()** the initial statement gets the information from the file stream object, which is passed as the first parameter, then creates a message to be given to the next component, writes the process of calling the next component, and completes the basic logic.
* * *
【Memo】For Studio, launch the Business Services Wizard in the New Creation menu, select the adapter and press the Finish button.
* * *
The **OnProcessInput()** method definition is as follows:
```objectscript
Method OnProcessInput(pInput As %Stream.Object, Output pOutput As %RegisteredObject) As %Status
```
**pInput** is provided with an instance of the **%Stream.FileCharacter** class for text files or the **%Stream.FileBinary** class for binary files.
In the sample, a file in text format will be inputted, and we have written it to accept multi-line requests and one request per line.
**AtEnd property** is set to 1 when EndOfFile is deteced. You can use this property to stop loop.
In a loop, we read the lines using the **ReadLine()** method, which enables us to obtain information about the contents of the file one line at a time (see the documentation for file adapter details).
Compose the message, retrieving information line by line. Then, we execute the ..SendRequestAsync() method, which calls the other components.
When executing the method, the first parameter should be the name of the component you want to call as a string, and the second parameter should be the request message you have created.
Note that the ..SendRequestAsync() is an asynchronous call and does not wait for a response.
Memo:There is also SendRequestSync() for synchronous calls.。
The sample code is as follows:
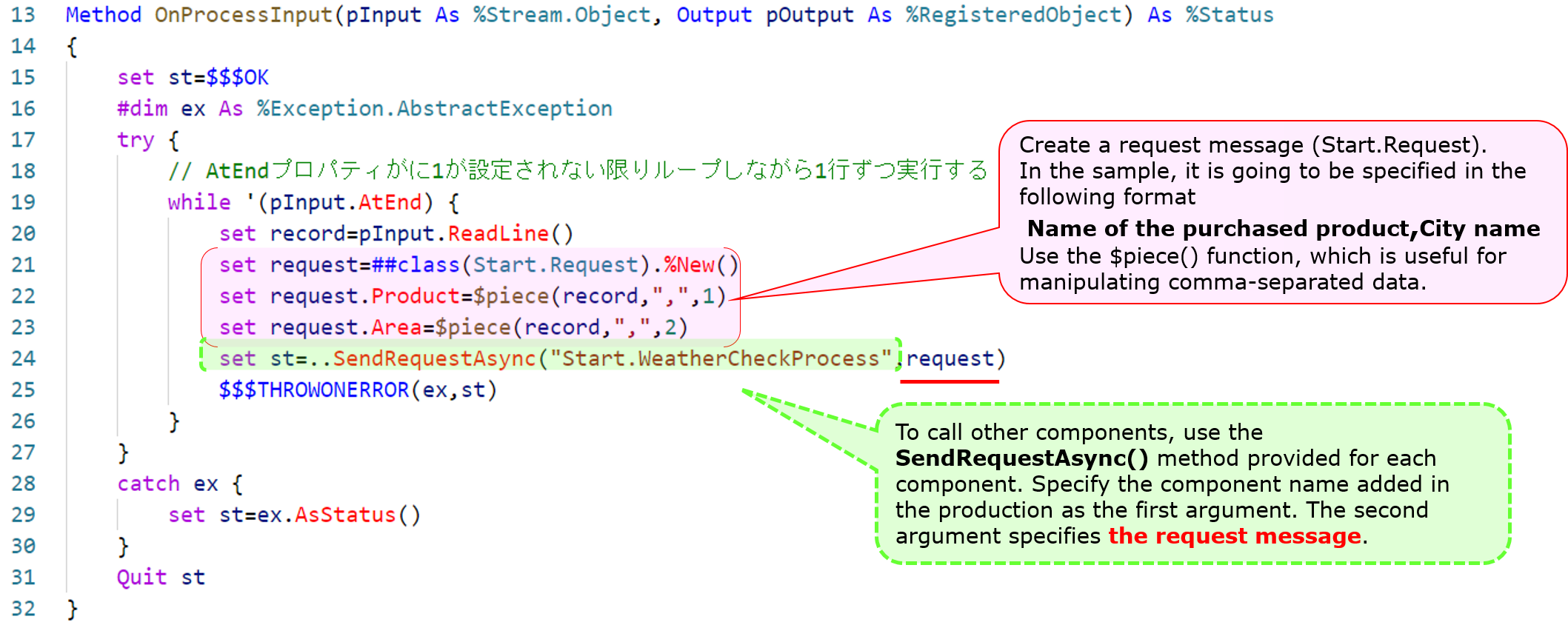
Reference:explanation of the usage of the $piece() function in the above example text
$piece(“string”, ”delimiter mark”, ”position number”)
The function to set/get a string with a delimiter, in the sample, to get the first and second value of comma-separated data, is written with the following syntax:
```objectscript
set request.Product=$piece(record,",",1)
set request.Area=$piece(record,",",2)
```
Now, let's check the function of Start.FileBS as it appeared in the above description.
In the sample production, the "File Path" was set to **/irisdev/src**, and the "File Spec" was set to **check.txt**. Either prepare the same directory or change it to a different directory and register the sample data in the check.txt file using the following format: purchased product name, name of the city.
※If you are using the sample container, please rename [Test-check.txt] in the src directory under the directory created by the git clone.
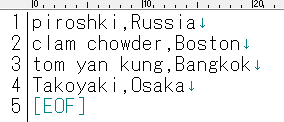
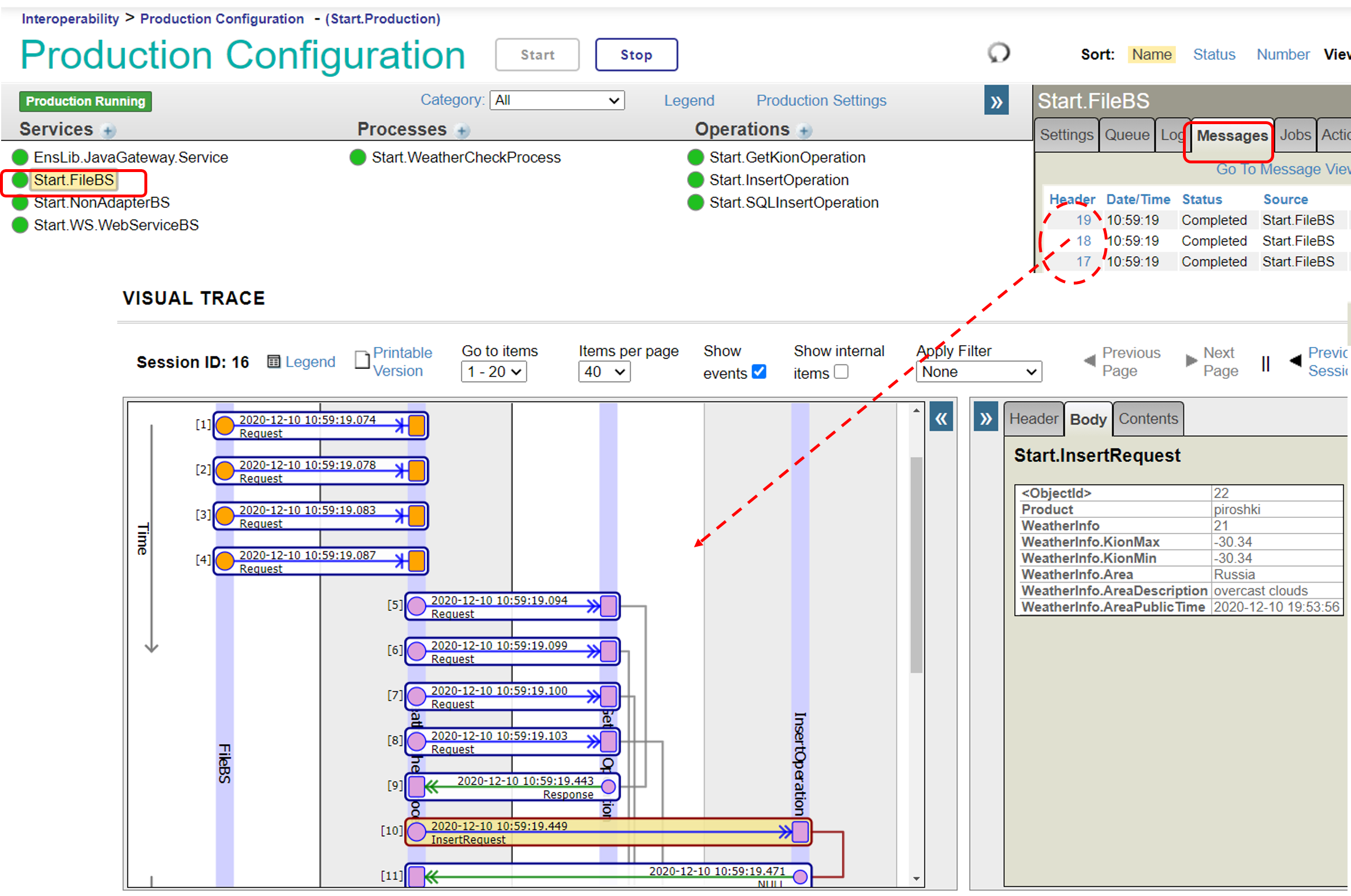
### 2. Business services for Web services using the SOAP inbound adapter (Start.WS.WebServiceBS)
Next, we will outline the creation of business services for Web services.
The Business Service Class for Web Services acts as a Web Service Provider = Web Service Server.
In the sample, we have two parameters in the Web service method for this sample production to have information sent from the Web service client. The web method uses the data entered in the parameters to create a message class and call other components.

When you define a Web service class, a test screen is created. However, it is not shown by default.
Log in to IRIS (or start a terminal), go to the namespace where the production is located and do the following:
For your reference:Access to the Catalog and Test Pages
Here is a sample code configuration in the setting where the container was started with docker-compose up -d (run in the %SYS namespace)
set $namespace="%SYS"
set ^SYS("Security","CSP","AllowClass","/csp/user/","%SOAP.WebServiceInfo")=1
set ^SYS("Security","CSP","AllowClass","/csp/user/","%SOAP.WebServiceInvoke")=1
【Attention】Please note that the sentence is case-sensitive and should be written with care. Also, depending on the namespace in which the product is used, the specified script changes. The example sentence is written on the assumption that the sample is imported into the USER namespace.If you import the sample code into the ABC namespace, the fourth subscript should be "/csp/abc/."
Once the configuration is complete, go to the following URL:
http://localhost:52773/csp/user/Start.WS.WebServiceBS.cls
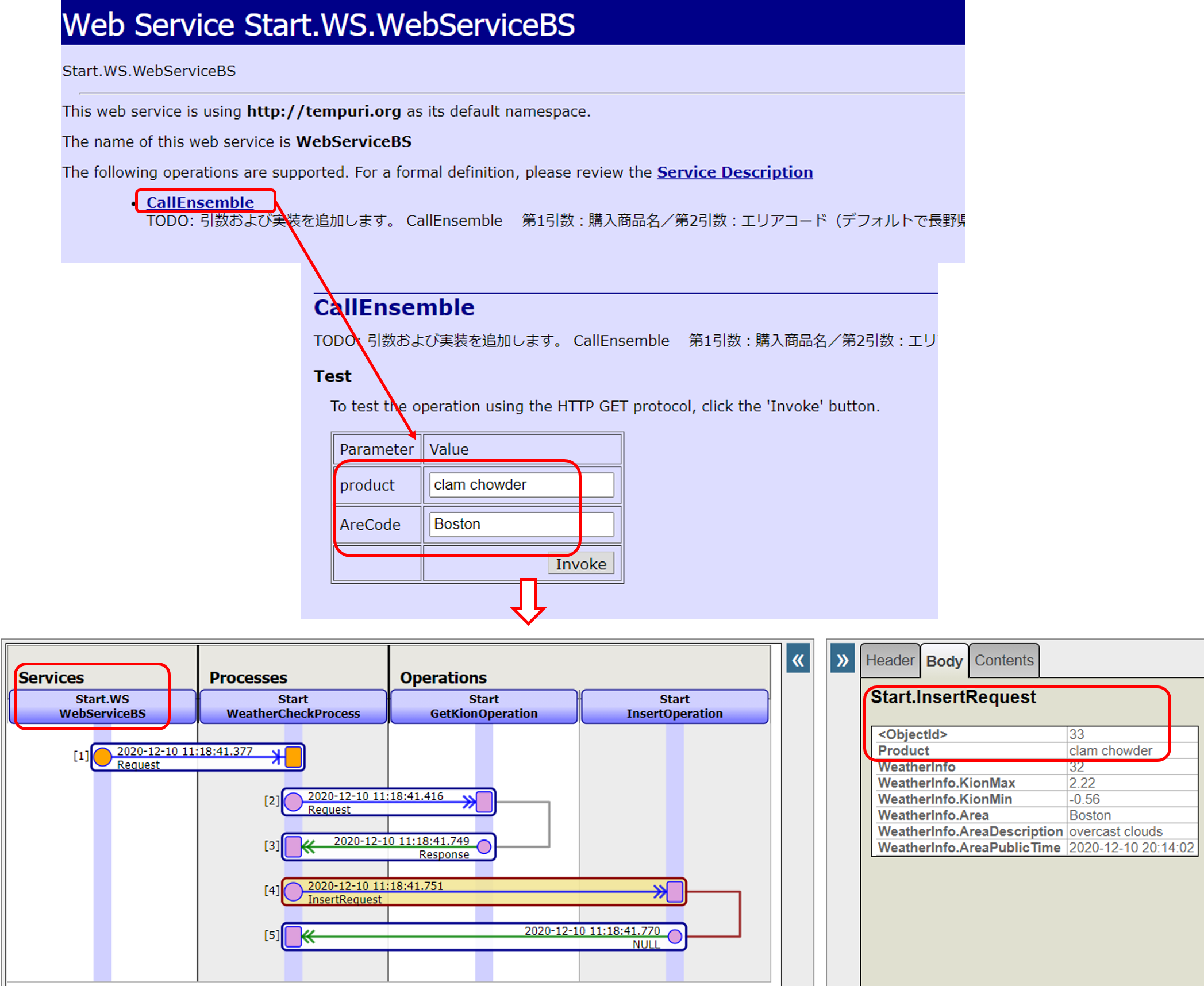
If you want to provide the WSDL to your Web services client, specify WSDL=1 at the end of the following URL
http://localhost:52773/csp/user/Start.WS.WebServiceBS.cls?WSDL
### 3. Business services called by stored procedures or REST without using adapters(Start.NonAdapterBS)
Next, we will introduce the Business Service without adapters (Start.NonAdapterBS).
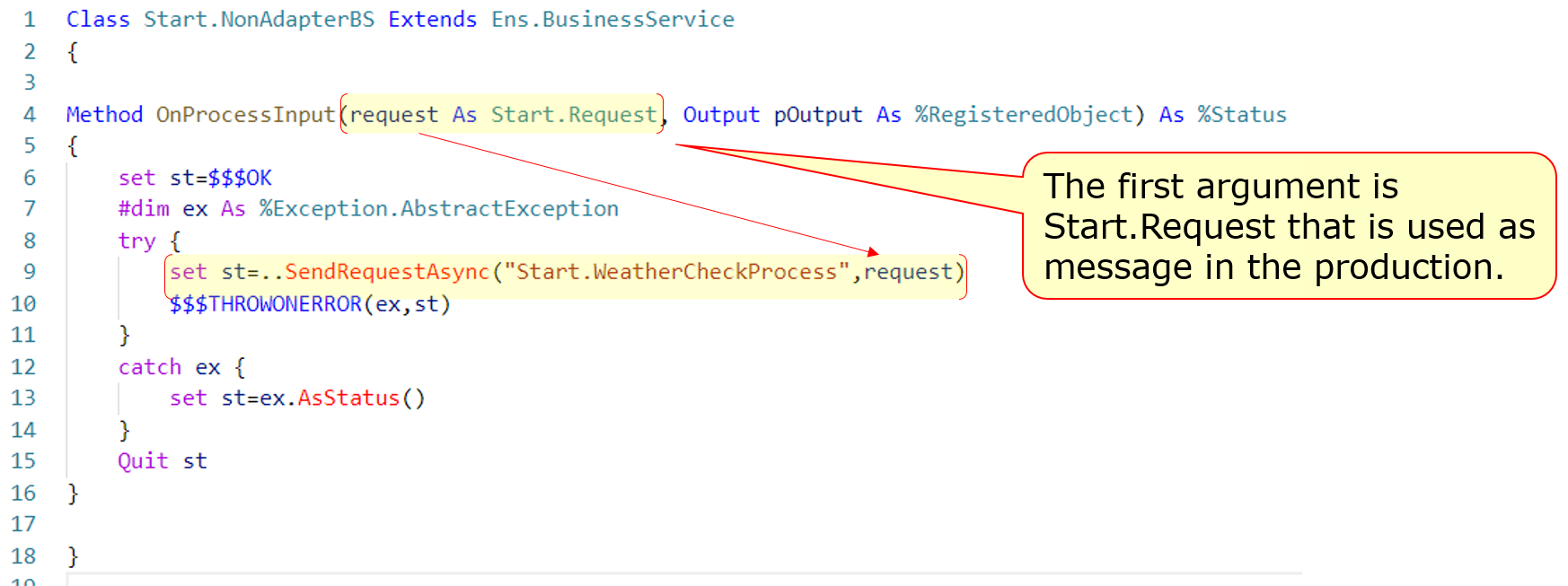
For business services that use adapters, the adapter calls the business service's ProcessInput() method to detect the information.
If you don't use adapters, you can still call the ProcessInput() method, but this method is not public. Therefore, if you implement a business service that does not use adapters, you will need to consider ProcessInput().
The sample utilizes the following two methods:
* Stored procedures(Start.Utils)
* Dispatch Class for REST(Start.REST)→This is the service we ran in this article.
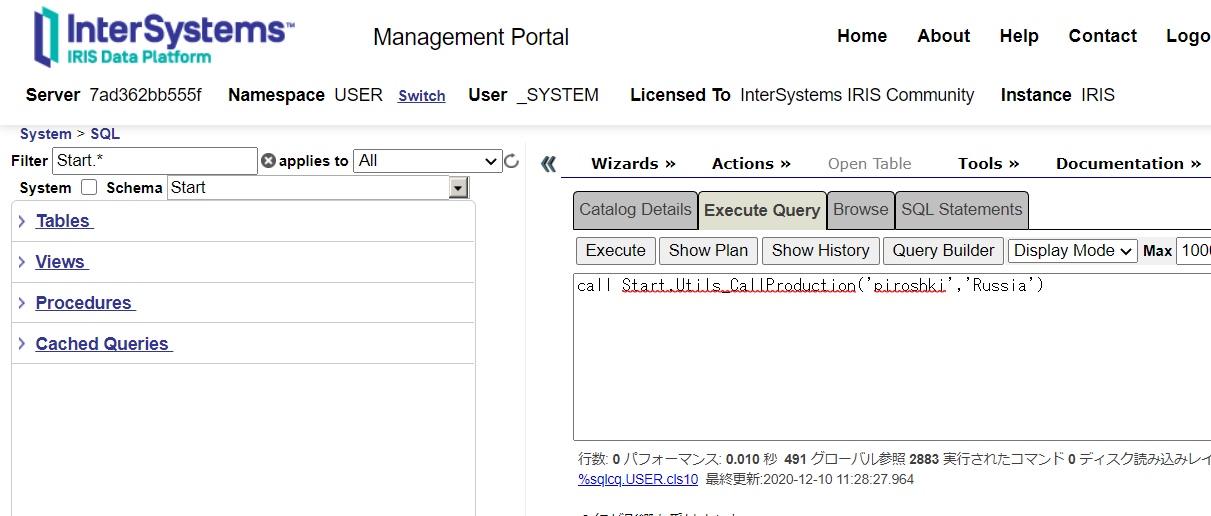
Now, here's an example of a stored procedure.
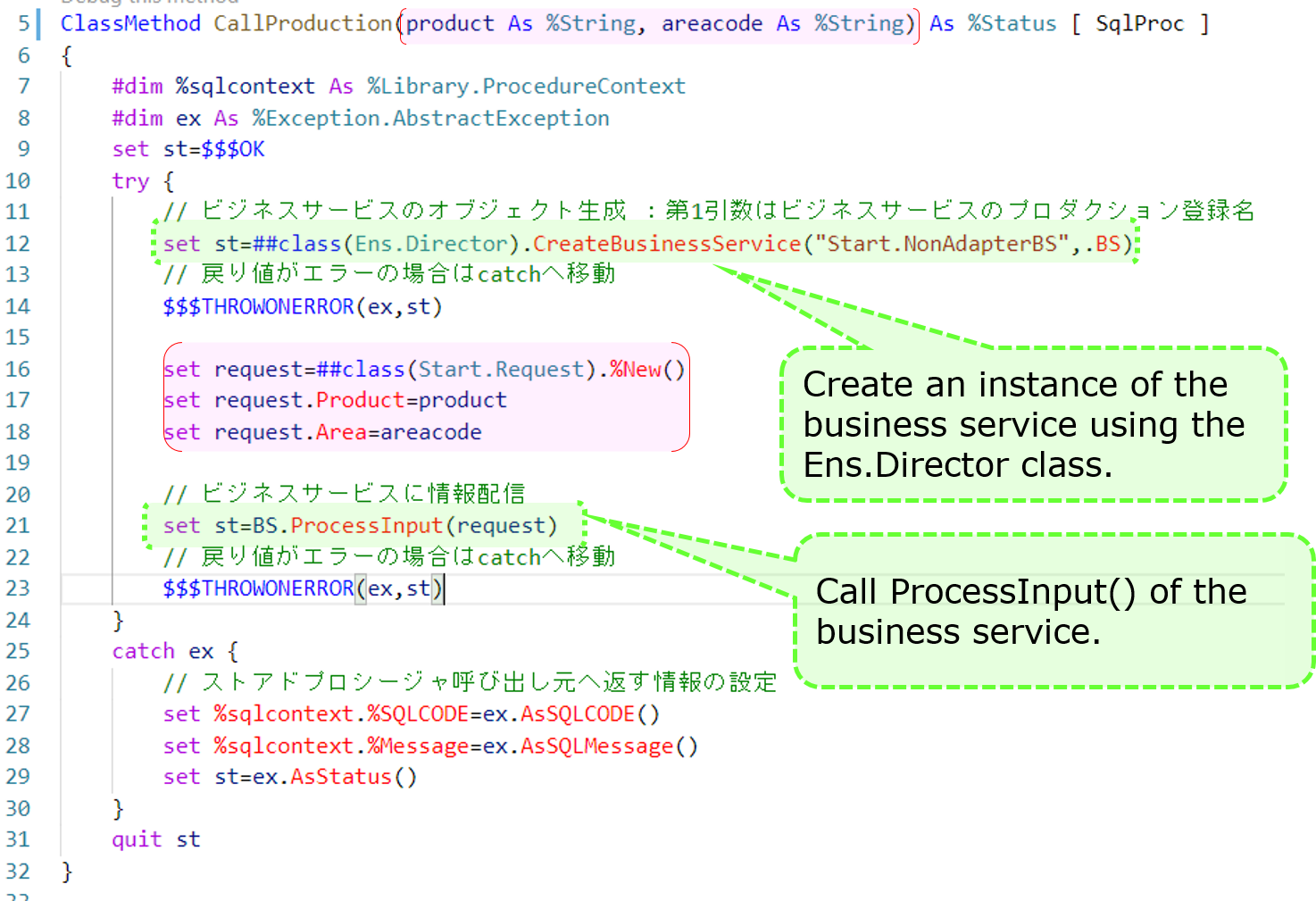
After adding a business service (Start.NonAdapterBS) that does not use adapters to the production (state added in the sample), run the following stored procedure
call Start.Utils_CallProduction('piroshki','Russia')
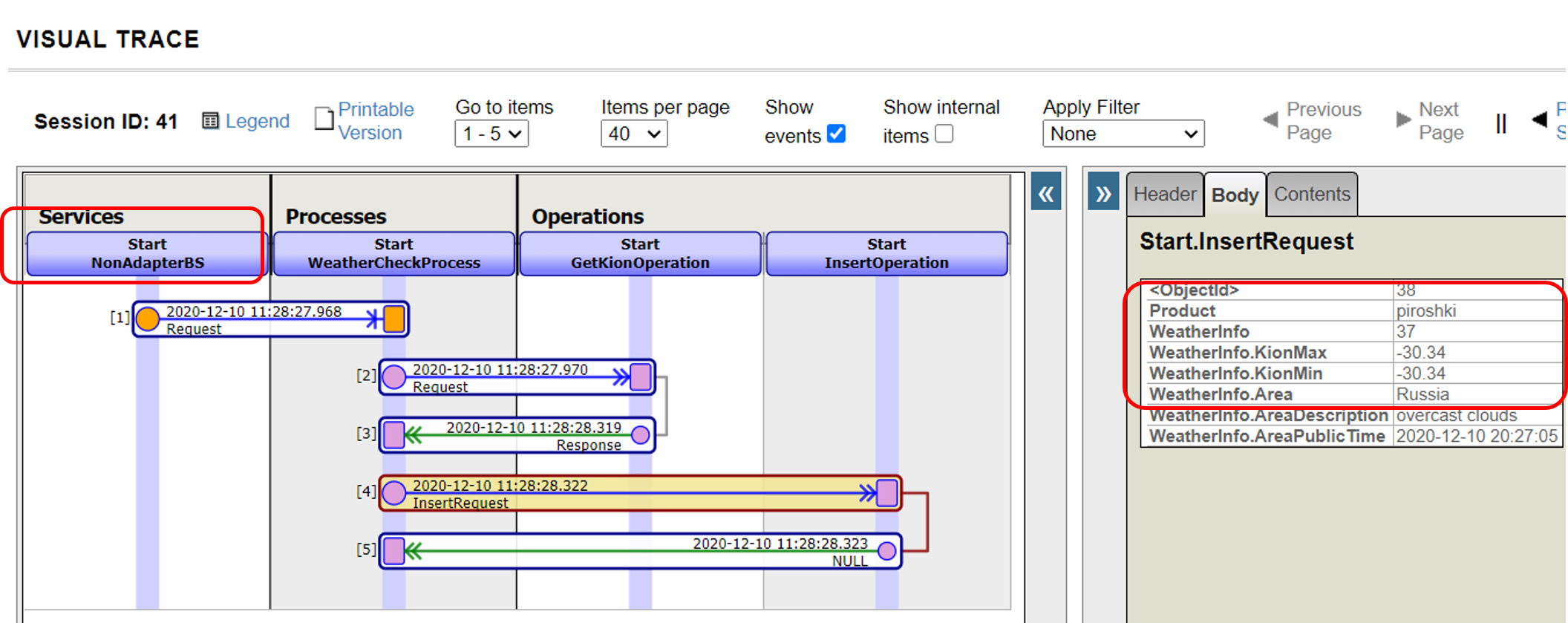
A resulting trace of the running result is as follows:
Next, here is an example of creating a dispatch class for REST:
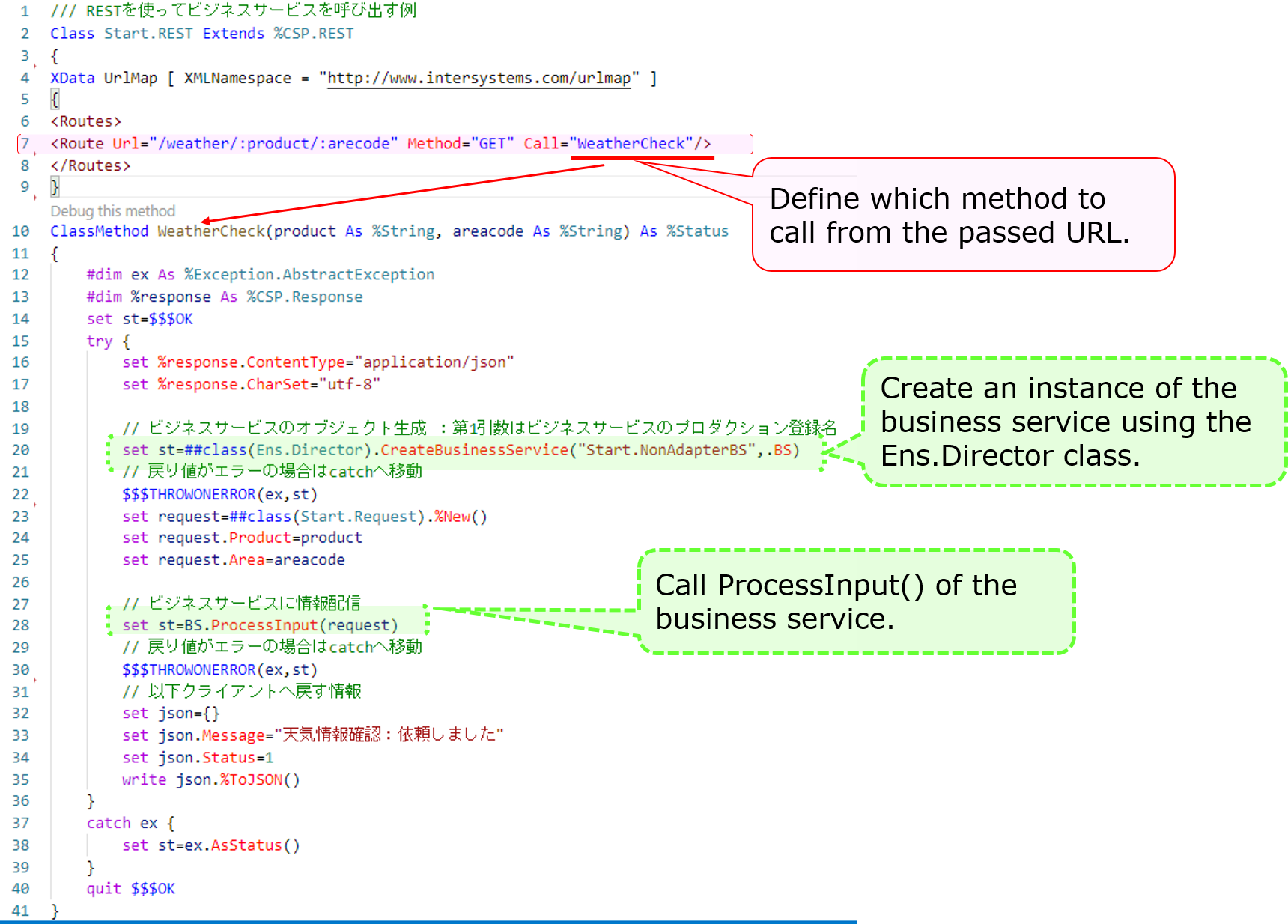
The XML described in the XData Url Map defines which methods are called in response to the URL at the time of the REST call.
The example describes a definition that calls the **WeatherCheck()** method when the URL of the **/weather/first parameter (purchased product name)/ second parameter (name of the city)** are provided in the **GET request**.
```objectscript
```
Then, define the base URL for the above URL in the Management Portal's Web Application Path Settings screen, and it is complete.
See this article for more details on the configuration.
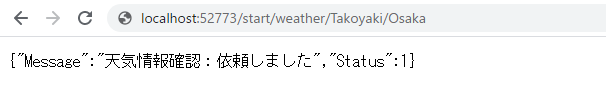
Once it is ready, try to run the information using a business service that allows you to send the REST information.
Example)http://localhost:52773/start/weather/Takoyaki/Osaka
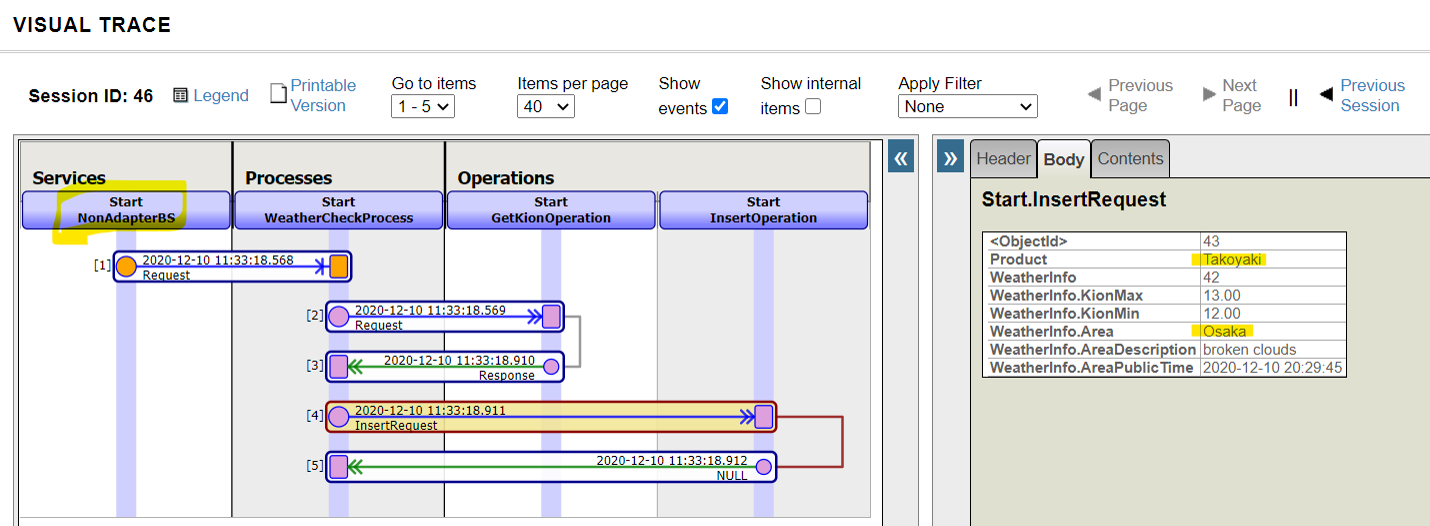
If you do not use an adapter, as ProcessInput() cannot be called directly from outside, we have created an object for the business service in the logic executed through REST or stored procedures (using the CreateBusinessService() method of the Ens.Director class) and called ProcessInput()
If you use an adapter, the adapter detects the input and stores the information in a unique object and passes it to the business service. In contrast, if you don't use an adapter, the rest is pretty much the same, only the difference is in the above-mentioned part of the process.
The business service is simply designed to use the information entered outside IRIS to create request messages and call business components.
Throughout the sample production, we were able to see the following:
Different components play different roles in making a production run (business services, business processes, business operations).
To transmit information between components, use the message.
Messages are stored in the database unless deleted and thus can be traced at any time.
Some adapters simplify the process of around the connection.
These are the basic operations on how to use Interoperability in IRIS.
There are also record maps (see: FAQ TOPIC) and data conversion tools that are useful for input and output of CSV files and other format-specific files.
As well as this series, there is also an article on simple IoT applications developed with InterSystems IRIS using Interoperability. Please check it out.
Besides, IRIS for Health also supports FHIR and HL7 (including SS-MIX2) transmissions.
I would be pleased to explain it in another post. If you have something of interest to share, please leave a comment!
Finally, training courses are also available to learn how to use Interoperability.
If you'd like to take the time to try it out with an instructor, please consider joining one of our training courses!
Announcement
Anastasia Dyubaylo · Mar 19, 2021
Hi Community,
Please welcome the new video on InterSystems Developers YouTube:
⏯ Deploying InterSystems IRIS Solutions into Kubernetes Google Cloud
See how an InterSystems IRIS data platform application is deployed into a Kubernetes cluster, specifically on Google Kubernetes Engine (GKE), using Terraform to create a cluster and a CI/CD GitHub implementation called GitHub Actions to automate deployment steps.
⬇️ Access all code samples here.
🗣 Presenter: @Mikhail.Khomenko, DevOps Engineer
Additional materials to this video you can find in this InterSystems Online Learning Course.
Enjoy watching this video! 👍🏼
Article
Evgeny Shvarov · Jun 24, 2020
Hi Developers!
Suppose you have a persistent class with data and you want to have a simple Angular UI for it to view the data and make CRUD operations.
Recently @Alberto.Fuentes described how to build Angular UI for your InterSystems IRIS application using RESTForms2.
In this article, I want to tell you how you can get a simple Angular UI to CRUD and view your InterSystems IRIS class data automatically in less than 5 minutes.
Let's go!
To make this happen you need:
1. InterSystems IRIS
2. ZPM
3. RESTForms2 and RESTForms2-UI modules.
I'll take a Data.Countries class which I generated and imported via csvgen using this command:
d ##class(community.csvgen).GenerateFromURL("https://raw.githubusercontent.com/datasciencedojo/datasets/master/WorldDBTables/CountryTable.csv",",","Data.Countries"
To make an Angular UI we need to expose REST API for this class, which will service CRUD operations.
Let's use restforms2 module for this.
This command in dockerfile installs restforms2 into IRIS container:
zpm "install restforms2" \
To add a REST API we need to derive the class from Form.Adaptor:
Class Data.Countries Extends (%Library.Persistent, Form.Adaptor)
Add restforms2 parameters to the persistent class to manage the general behavior: sorting parameter, display name, etc:
// Form name, not a global key so it can be anything
Parameter FORMNAME = "Countries";
/// Default permissions
/// Objects of this form can be Created, Read, Updated and Deleted
/// Redefine this parameter to change permissions for everyone
/// Redefine checkPermission method (see Form.Security) for this class
/// to add custom security based on user/roles/etc.
Parameter OBJPERMISSIONS As %String = "CRUD";
/// Property used for basic information about the object
/// By default getObjectDisplayName method gets its value from it
Parameter DISPLAYPROPERTY As %String = "name";
Perfect. Next, we can use restforms2 syntax to let restforms2 know, what properties of the class we want to expose to the CRUD. You can make it adding "DISPLAYNAME =" attribute to the properties, you want to expose into restforms2-ui. Example:
Property code As %Library.String(MAXLEN = 250) [ SqlColumnNumber = 2 ];
Property name As %Library.String(DISPLAYNAME = "Name", MAXLEN = 250) [ SqlColumnNumber = 3 ];
Property continent As %Library.String(DISPLAYNAME = "Continent", MAXLEN = 250) [ SqlColumnNumber = 4 ];
Property region As %Library.String(DISPLAYNAME = "Region", MAXLEN = 250) [ SqlColumnNumber = 5 ];
Property surfacearea As %Library.Integer(DISPLAYNAME = "Surface Area", MAXVAL = 2147483647, MINVAL = -2147483648) [ SqlColumnNumber = 6, SqlFieldName = surface_area ];
Property independenceyear As %Library.Integer(DISPLAYNAME = "Independence Year", MAXVAL = 2147483647, MINVAL = -2147483648) [ SqlColumnNumber = 7, SqlFieldName = independence_year ];
Great! Now lets introduce the UI layer. This command in dockerfile installs restforms2-ui, which is Angular UI for Restform2:
zpm "install restforms2-ui" \
That's it! Let' examine the UI for your class, which you can find in the URL server:port/restforms2-ui:
RESTForms goes with test classes Person and Company - and you can use it to examine the features of restformsUI. Currently It can edit string, number, boolean, date and look-up fields.
You can test all this on your laptop, if clone and build this repository:
docker-compose up -d --build
And then open the URL:
localhost:port/restforms2-ui/index.html
or if you use VSCode, select this menu item:
Happy coding and stay tuned! It's great! I tried the application, and I liked the interface and how easy it is to create a simple CRUD using RESTForms. 💡 This article is considered as InterSystems Data Platform Best Practice. Love the accelerator concept for quick and easy CRUD :)
Announcement
Anastasia Dyubaylo · Sep 29, 2020
Hey Developers,
New demo show by InterSystems Manager @Amir.Samary is already on InterSystems Developers YouTube:
⏯ InterSystems IRIS: Kafka, Schema Normalization and Service Enablement
Demo of InterSystems IRIS with Kafka, Schema Registry, AVRO and Schema Migration.
This demo allows you to show:
A Bank Simulator generating AVRO messages and sending them over Kafka to InterSystems IRIS
InterSystems IRIS Multi-model capabilities (AVRO, JSON, Objects, SQL and MDX)
How InterSystems IRIS can transform the AVRO object into a canonical structure using transformations and lookups
How InterSystems IRIS can orchestrate this work and start a human workflow in case of a problem during the transformations
How InterSystems IRIS can provide bidirectional data lineage (from source to canonical and vice-versa)
How InterSystems IRIS pull new schemas from a Kafka Schema Registry and generate the data structures automatically to support schema evolution
This demo uses Confluent's Kafka and their docker-compose sample
⬇️ Kafka Demo with InterSystems IRIS on Open Exchange
This demo can be also found at https://github.com/intersystems-community/irisdemo-demo-kafka
Enjoy watching the video! 👍🏼
Announcement
Olga Zavrazhnova · Feb 26, 2021
Hi Developers,A new exciting challenge introduced for Global Masters members of "Advocate" level and above: we invite you to record a 30-60 sec video with an answer to our question:
➥ What is the value of InterSystems IRIS to you?
🎁 Reward of your choice for doing the interview: $50 Gift Card (VISA/Amazon) or 12,000 points!
Follow this direct link to the challenge for more information. Please note that the link will work for GM members of "Advocate" level and above. More about GM levels you can read here.
We would love to hear from you!
See you on the Global Masters Advocate Hub today!
Announcement
Stefan Wittmann · Jul 1, 2020
GA releases are now published for the 2020.2 version of InterSystems IRIS, IRIS for Health, and IRIS Studio!
A full set of containers for these products are available from the WRC Software Distribution site, including community editions of InterSystems IRIS and IRIS for Health.
The build number for these releases is 2020.2.0.211.0.
InterSystems IRIS Data Platform 2020.2 provides an important security update with the following enhancements:
Support for TLS 1.3
Support for SHA-3
InterSystems IRIS for Health 2020.2 includes all of the enhancements of InterSystems IRIS. In addition, this release includes:
FHIR R4 Data transformations
New configuration UI for the FHIR Server
Support for IHE RMU Profile
IHE Connectathon Updates
As this is a CD (Continuous Delivery) release, many customers may want to know the differences between 2020.2 and 2020.1. These are listed in the release notes:
InterSystems IRIS 2020.2 release notes
IRIS for Health 2020.2 release notes
Documentation can be found here:
InterSystems IRIS 2020.2 documentation
IRIS for Health 2020.2 documentation
InterSystems IRIS Studio 2020.2 is a standalone development image supported on Microsoft Windows. It works with InterSystems IRIS and IRIS for Health version 2020.2 and below, as well as with Caché and Ensemble.
The platforms on which InterSystems IRIS and IRIS for Health 2020.2 are supported for production and development are detailed in the Supported Platforms document. Hi Stefan,
I'm not seeing the 2020.2 distributions listed for the full kits on the WRC site. Do you know when they will be available? Hi @Jeffrey.Drumm ,
thanks for the catch, the wording was not clear and I have corrected the statement. 2020.2 is a CD release, which only contains container images, not full kits.
Thanks,
Stefan Full kits will not be available for this release? Correct. @Jeffrey.Drumm - please see the article on how InterSystems IRIS will be released:
https://community.intersystems.com/post/new-release-cadence-intersystems-irisThis may help to clear up some confusion. The Community Edition images for IRIS and IRIS for Health are now available in the Docker Store. And we now offer the Community Edition for both x64 and ARM64 architectures. Try them out!
InterSystems IRIS:
docker pull store/intersystems/iris-community:2020.2.0.211.0
docker pull store/intersystems/iris-community-arm64:2020.2.0.211.0
InterSystems IRIS for Health:
docker pull store/intersystems/irishealth-community:2020.2.0.211.0
docker pull store/intersystems/irishealth-community-arm64:2020.2.0.211.0 Thank you for the clarification. I'm in the middle of a Cache/Ensemble on SUSE -> IRIS for Health on RHEL migration and would prefer to be at the latest available release prior to go-live. But I guess I am, per the release schedule. Great news about ARM support.
As tagged image can be provided with several architectures (example with 4) is there a reason why we separate the tags into community and community-arm? As I understand, this functionality (as well as the ability to use default image tags such as 'openjdk' vs '/store/intersystems/...') is only available for Official Images (https://docs.docker.com/docker-hub/official_images/) that are published by Docker. We are a verified publisher, and our images are Docker certified, but they are not published and maintained by Docker. Here's a Docker Hub image (not official, not certified) with multiple architectures.
Here's info on publishing multi-arch images. Some more info.
manifest command was successfully merged into docker codebase so it seems possible now. Are there any plans to bring the FHIR capabilities to the EM channel - 2020.1.x? While I appreciate the difference between CD/EM models, these FHIR updates could be critical for those of us that have to stick to the EM model (particularly the FHIR R4 and FHIR Server changes) without needing to wait until a 2021.x release. There are no plans to integrate latest FHIR features into EM (Extended Maintenance) releases at this time, but we will be sure to let everyone know should our release plans change in the future.
Thanks,Craig
Article
Eduard Lebedyuk · Aug 3, 2020
InterSystems IRIS currently limits classes to 999 properties.
But what to do if you need to store more data per object?
This article would answer this question (with the additional cameo of Community Python Gateway and how you can transfer wide datasets into Python).
The answer is very simple actually - InterSystems IRIS currently limits classes to 999 properties, but not to 999 primitives. The property in InterSystems IRIS can be an object with 999 properties and so on - the limit can be easily disregarded.
Approach 1.
Store 100 properties per serial property. First create a stored class that stores a hundred properties.
Class Test.Serial Extends %SerialObject
{
Property col0;
...
Property col99;
}
And in your main class add as much properties as you need:
Class Test.Record Extends %Persistent
{
Property col00 As Test.Serial;
Property col01 As Test.Serial;
...
Property col63 As Test.Serial;
}
This immediately raises your limit to 99900 properties.
This approach offers uniform access for all properties via SQL and object layers (we always know property reference by it's number).
Approach 2.
One $lb property.
Class Test.Record Extends %Persistent
{
Property col As %List;
}
This approach is simpler but does not provide explicit column names.
Use SQL $LIST* Functions to access list elements.
Approach 3.
Use Collection (List Of/Array Of) property.
Class Test.Record Extends %Persistent
{
Property col As List Of %Integer;
}
This approach also does not provide explicit column names for individual values (but do you really need it?). Use property parameters to project the property as SQL column/table.
Docs for collection properties.
Approach 4.
Do not create properties at all and expose them via SQL Stored procedure/%DispatchGetProperty.
Class Test.Record Extends %Persistent
{
Parameter GLVN = {..GLVN("Test.Record")};
/// SELECT Test_Record.col(ID, 123)
/// FROM Test.Record
///
/// w ##class(Test.Record).col(1, )
ClassMethod col(id, num) As %Decimal [ SqlProc ]
{
#define GLVN(%class) ##Expression(##class(Test.Record).GLVN(%class))
quit $lg($$$GLVN("Test.Record")(id), num + 1)
}
/// Refer to properties as: obj.col123
Method %DispatchGetProperty(Property As %String) [ CodeMode = expression ]
{
..col(..%Id(), $e(Property, 4, *))
}
/// Get data global
/// w ##class(Test.Record).GLVN("Test.Record")
ClassMethod GLVN(class As %Dictionary.CacheClassname = {$classname()}) As %String
{
return:'$$$comClassDefined(class) ""
set strategy = $$$comClassKeyGet(class, $$$cCLASSstoragestrategy)
return $$$defMemberKeyGet(class, $$$cCLASSstorage, strategy, $$$cSDEFdatalocation)
}
The trick here is to store everything in the main $lb and use unallocated schema storage spaces to store your data. Here's an article on global storage.
With this approach, you can also easily transfer the data into Python environment with Community Python Gateway via the ExecuteGlobal method.
This is also the fastest way to import CSV files due to the similarity of the structures.
Conclusion
999 property limit can be easily extended in InterSystems IRIS.
Do you know other approaches to storing wide datasets? If so, please share them!
The question is how csvgen could be upgraded to consume csv files with 1000+ cols. While I always advertise CSV2CLASS methods for generic solutions, wide datasets often possess an (un)fortunate characteristic of also being long.
In that case custom object-less parser works better.
Here's how it can be implemented.
1. Align storage schema with CSV structure
2. Modify this snippet for your class/CSV file:
Parameter GLVN = {..GLVN("Test.Record")};
Parameter SEPARATOR = ";";
ClassMethod Import(file = "source.csv", killExtent As %Boolean = {$$$YES})
{
set stream = ##class(%Stream.FileCharacter).%New()
do stream.LinkToFile(file)
kill:killExtent @..#GLVN
set i=0
set start = $zh
while 'stream.AtEnd {
set i = i + 1
set line = stream.ReadLine($$$MaxStringLength)
set @..#GLVN($i(@..#GLVN)) = ..ProcessLine(line)
write:'(i#100000) "Processed:", i, !
}
set end = $zh
write "Done",!
write "Time: ", end - start, !
}
ClassMethod ProcessLine(line As %String) As %List
{
set list = $lfs(line, ..#SEPARATOR)
set list2 = ""
set ptr=0
// NULLs and numbers handling.
// Add generic handlers here.
// For example translate "N/A" value into $lb() if that's how source data rolls
while $listnext(list, ptr, value) {
set list2 = list2 _ $select($g(value)="":$lb(), $ISVALIDNUM(value):$lb(+value), 1:$lb(value))
}
// Add specific handlers here
// For example convert date into horolog in column4
// Add %%CLASSNAME
set list2 = $lb() _ list2
quit list2
} Thanks, Ed!Could you make a PR? I have no concrete ideas on how to automate this.
This is a more case-by-case basis. After more than 42 years of M-programming and in total of 48 years of programming experience I would say, if you need a class with about 1000 or more properties than something is wrong with your (database) design. There is nothing more to say. Period. Wide datasets are fairly typical for:
Industrial data
IoT
Sensors data
Mining and processing data
Spectrometry data
Analytical data
Most datasets after one-hot-encoding applied
NLP datasets
Any dataset where we need to raise dimensionality
Media featuresets
Social Network/modelling schemas
I'm fairly sure there's more areas but I have not encountered them myself.
Recently I have delivered a PoC with classes more than 6400 columns wide and that's where I got my inspiration for this article (I chose approach 4).
@Renato.Banzai also wrote an excellent article on his project with more than 999 properties.
Overall I'd like to say that a class with more than 999 properties is a correct design in many cases. You probably right for a majority of tasks. But how do you manage with AI tasks which NEED to manage thousands of features of entities? And features are properties/fields from data storage perspective.
Anyway, I'm really curious how do you deal with AI/ML tasks in IRIS or Caché. Entity–attribute–value model is usually used for this purpose.
I have already written about this at the time: SQL index for array property elements. That's good and well for sparse datasets (where say you have a record with 10 000 possible attributes but on average only 50 are filled).
EAV does not help in dense cases where every record actually has 10 000 attributes. My EAV implementation is the same as your Approach 3, so it will work fine even with fully filled 4.000.000 attributes.
Since the string has a limit of 3,641,144, approaches with serial and %List are dropped.
All other things being equal, everything depends on the specific technical task: speed, support for Objects/SQL, the ability to name each attribute, the number of attributes, and so on. The approach 1 doesn't "raises your limit to 99900 properties" but rather to 6600 properties.
You can test it through large.utils.cls
w ##class(large.utils).init(66,100) deleting large.serial100creating large.serial100compiling large.serial100
Compilation started on 07/31/2023 14:48:16Compiling class large.serial100Compiling routine large.serial100.1Compilation finished successfully in 0.218s.
creating large.c66compiling large.c66
Compilation started on 07/31/2023 14:48:16Compiling class large.c66Compiling table large.c66Compiling routine large.c66.1Compilation finished successfully in 8.356s.
1w ##class(large.utils).init(67,100)deleting large.serial100creating large.serial100compiling large.serial100
Compilation started on 07/31/2023 14:48:27Compiling class large.serial100Compiling routine large.serial100.1Compilation finished successfully in 0.213s.
creating large.c67compiling large.c67
Compilation started on 07/31/2023 14:48:27Compiling class large.c67Compiling table large.c67Compiling routine large.c67.1ERROR #5002: ObjectScript error: <MAXSTRING>CompileRtns+286^%occRoutineERROR #5002: ObjectScript error: <NOROUTINE>DescribePhase2+9^%occSysDescriptor *large.c67.1Detected 2 errors during compilation in 6.896s.
0 a(<MAXSTRING>CompileRtns+286^%occRoutineÛCompileRtns+286^%occRoutine IRISAPP³e^ReturnError+2^%occSystem^1!e^CompileList+229^%occCompile^1e^CompileList+23^%apiOBJ^1e^Compile+1^%apiOBJ^1e^Compile+1^%SYSTEM.OBJ.1^1^init+50^large.utils.1^1e^^^0K0 G¦ large.c67/ IRISAPP#!e^CompileRtns+388^%occRoutine^1IRISAPP> I guess we are hitting some other limit.
I went with approach 4 myself. I fully agree with Julius.Any class that exceeds 100 properties is dead wrong.We're not talking tables (relational) (and even then it's wrong) but OO design, which doesn't seem to be much in fashion these days.Redesign your solution