Record linkage using InterSystems IRIS, Apache Zeppelin, and Apache Spark
Hi all. We are going to find duplicates in a dataset using Apache Spark Machine Learning algorithms.
Note: I have done the following on Ubuntu 18.04, Python 3.6.5, Zeppelin 0.8.0, Spark 2.1.1
Introduction
In previous articles we have done the following:
- The way to launch Jupyter Notebook + Apache Spark + InterSystems IRIS
- Load a ML model into InterSystems IRIS
- K-Means clustering of the Iris Dataset
- The way to launch Apache Spark + Apache Zeppelin + InterSystems IRIS
In this series of articles, we explore Machine Learning and record linkage.
Imagine that we merged databases of neighboring shops. Most probably there will be records that are very similar to each over. Some records will be of the same person and we call them duplicates. Our purpose is to find duplicates.
Why is this necessary? First of all, to combine data from many different operational source systems into one logical data model, which can then be subsequently fed into a business intelligence system for reporting and analytics. Secondly, to reduce data storage costs. There are some additional use cases.
Approach
What data do we have? Each row contains different anonymized information about one person. There are family names, given names, middle names, date of births, several documents, etc.
The first step is to look at the number of records because we are going to make pairs. The number of pairs equals n*(n-1)/2. So, if you have less than 5000 records, than the number of pairs would be 12.497.500. It is not that many, so we can pair each record. But if you have 50.000, 100.000 or more, the number of pairs more than a billion. This number of pairs is hard to store and work with.
So, if you have a lot of records, it would be a good idea to reduce this number. We will do it by selecting potential duplicates. A potential duplicate is a pair, that might be a duplicate. We will detect them based on several simple conditions. A specific condition might be like:
(record1.family_name == record2.familyName) & (record1.givenName == record2.givenName) & (record1.dateOfBirth == record2.dateOfBirth)
but keep in mind that you can miss duplicates because of strict logical conditions. I think the optimal solution is to choose important conditions and use no more than two of them with & operator. But you should convert each feature into one record shape beforehand. For example, there are several ways to store dates: 1985-10-10, 10/10/1985, etc convert to 10-10-1985(month-day-year).
The next step is to label the part of the dataset. We will randomly choose, for example, 5000-10000 pairs (or more, if you are sure that you can label all of them). We will save them to IRIS and label these pairs in Jupyter (Unfortunately, I didn't find an easy and convenient way to do it. Also, you can label them in PySpark console or wherever you want).
After that, we will make a feature vector for each pair. During the labeling process probably you noticed which features are important and what they equal. So, test different approaches to creating feature vectors.
Test different machine learning models. I chose a random forest model because of tests (accuracy/precision/recall/etc). Also, you can try decision trees, Naive Bayes, other classification model and choose the one that will be the best.
Test the result. If you are not satisfied with the result, try to change feature vectors or change a ML model.
Finally, fit all pairs into the model and look at the result.
Implementation
Load a dataset:
%pyspark
dataFrame=spark.read.format("com.intersystems.spark").option("url", "IRIS://localhost:51773/******").option("user", "*******").option("password", "*********************").option("dbtable", "**************").load()

Clean the dataset. For example, null (check every row) or useless columns:
%pyspark
columns_to_drop = ['allIdentityDocuments', 'birthCertificate_docSource', 'birthCertificate_expirationDate', 'identityDocument_expirationDate', 'fullName']
droppedDF = dataFrame.drop(*columns_to_drop)
Prepare the dataset for making pairs:
%pyspark
from pyspark.sql.functions import col
# rename columns names
replacements1 = {c : c + '1' for c in droppedDF.columns}
df1 = droppedDF.select([col(c).alias(replacements1.get(c, c)) for c in droppedDF.columns])
replacements2 = {c : c + '2' for c in droppedDF.columns}
df2 = droppedDF.select([col(c).alias(replacements2.get(c, c)) for c in droppedDF.columns])
To make pairs we will use join function with several conditions.
%pyspark
testTable = (df1.join(df2, (df1.ID1 < df2.ID2) & ((df1.familyName1 == df2.familyName2) & (df1.givenName1 == df2.givenName2)
| (df1.familyName1 == df2.familyName2) & (df1.middleName1 == df2.middleName2)
| (df1.familyName1 == df2.familyName2) & (df1.dob1 == df2.dob2)
| (df1.familyName1 == df2.familyName2) & (df1.snils1 == df2.snils2)
| (df1.familyName1 == df2.familyName2) & (df1.addr_addressLine1 == df2.addr_addressLine2)
| (df1.familyName1 == df2.familyName2) & (df1.addr_okato1 == df2.addr_okato2)
| (df1.givenName1 == df2.givenName2) & (df1.middleName1 == df2.middleName2)
| (df1.givenName1 == df2.givenName2) & (df1.dob1 == df2.dob2)
| (df1.givenName1 == df2.givenName2) & (df1.snils1 == df2.snils2)
| (df1.givenName1 == df2.givenName2) & (df1.addr_addressLine1 == df2.addr_addressLine2)
| (df1.givenName1 == df2.givenName2) & (df1.addr_okato1 == df2.addr_okato2)
| (df1.middleName1 == df2.middleName2) & (df1.dob1 == df2.dob2)
| (df1.middleName1 == df2.middleName2) & (df1.snils1 == df2.snils2)
| (df1.middleName1 == df2.middleName2) & (df1.addr_addressLine1 == df2.addr_addressLine2)
| (df1.middleName1 == df2.middleName2) & (df1.addr_okato1 == df2.addr_okato2)
| (df1.dob1 == df2.dob2) & (df1.snils1 == df2.snils2)
| (df1.dob1 == df2.dob2) & (df1.addr_addressLine1 == df2.addr_addressLine2)
| (df1.dob1 == df2.dob2) & (df1.addr_okato1 == df2.addr_okato2)
| (df1.snils1 == df2.snils2) & (df1.addr_addressLine1 == df2.addr_addressLine2)
| (df1.snils1 == df2.snils2) & (df1.addr_okato1 == df2.addr_okato2)
| (df1.addr_addressLine1 == df2.addr_addressLine2) & (df1.addr_okato1 == df2.addr_okato2))))
Check the size of returned dataframe:
%pyspark
droppedColumns = ['prevIdentityDocuments1', 'birthCertificate_docDate1', 'birthCertificate_docNum1', 'birthCertificate_docSer1', 'birthCertificate_docType1', 'identityDocument_docDate1', 'identityDocument_docNum1', 'identityDocument_docSer1', 'identityDocument_docSource1', 'identityDocument_docType1', 'prevIdentityDocuments2', 'birthCertificate_docDate2', 'birthCertificate_docNum2', 'birthCertificate_docSer2', 'birthCertificate_docType2', 'identityDocument_docDate2', 'identityDocument_docNum2', 'identityDocument_docSer2', 'identityDocument_docSource2', 'identityDocument_docType2']print(testTable.count())
testTable.drop(*droppedColumns).show() # I dropped several columns just for show() function
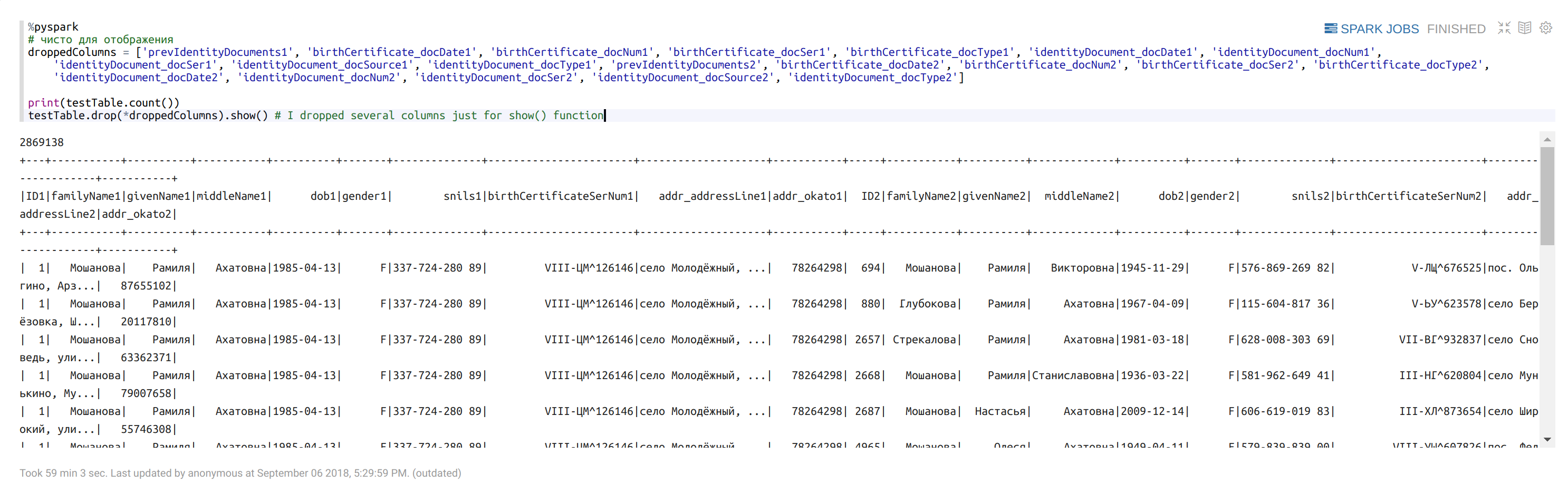
Randomly take a part of the dataframe:
%pyspark
randomDF = testTable.sample(False, 0.33, 0)
randomDF.write.format("com.intersystems.spark").\
option("url", "IRIS://localhost:51773/DEDUPL").\
option("user", "*****").option("password", "***********").\
option("dbtable", "deduplication.unlabeledData").save()
Label pairs in Jupyter
Run the following (it will widen the cells).
from IPython.core.display import display, HTML
display(HTML("<style>.container { width:100% !important; border-left-width: 1px !important; resize: vertical}</style>"))
Load dataframe:
unlabeledDF = spark.read.format("com.intersystems.spark").option("url", "IRIS://localhost:51773/DEDUPL").option("user", "********").option("password", "**************").option("dbtable", "deduplication.unlabeledData").load()
Return all the elements of the dataset as a list:
rows = labelledDF.collect()
The convenient way to display pairs:
from IPython.display import clear_output
from prettytable import PrettyTable
from collections import OrderedDictdef printTable(row):
row = OrderedDict((k, row.asDict()[k]) for k in newColumns)
table = PrettyTable()
column_names = ['Person1', 'Person2']
column1 = []
column2 = []
i = 0
for key, value in row.items():
if key != 'ID1' and key != 'ID2' and key != "prevIdentityDocuments1" and key != 'prevIdentityDocuments2' and key != "features":
if (i < 20):
column1.append(value)
else:
column2.append(value)
i += 1
table.add_column(column_names[0], column1)
table.add_column(column_names[1], column2)
print(table)
List where we will store rows:
listDF = []
The labeling process:
from pyspark.sql import Row
from IPython.display import clear_output
import time
# 3000 - 4020
for number in range(3000 + len(listDF), len(rows)):
row = rows[number]
if (len(listDF) % 10) == 0:
print(3000 + len(listDF))
printTable(row)
result = 0
label = 123
while True:
result = input('duplicate? y|n|stop')
if (result == 'stop'):
break
elif result == 'y':
label = 1.0
break
elif result == 'n':
label = 0.0
break
else:
print('only y|n|stop')
continue
if result == 'stop':
break
tmp = row.asDict()
tmp['label'] = label
newRow = Row(**tmp)
listDF.append(newRow)
time.sleep(0.2)
clear_output()

Create a dataframe again:
newColumns.append('label')
labelledDF = spark.createDataFrame(listDF).select(*newColumns)
Save it to IRIS:
labeledDF.write.format("com.intersystems.spark").\
option("url", "IRIS://localhost:51773/DEDUPL").\
option("user", "***********").option("password", "**********").\
option("dbtable", "deduplication.labeledData").save()
Feature vector and ML model
Load a dataframe into Zeppelin:
%pyspark
labeledDF = spark.read.format("com.intersystems.spark").option("url", "IRIS://localhost:51773/DEDUPL").option("user", "********").option("password", "***********").option("dbtable", "deduplication.labeledData").load()
Feature vector generation:
%pyspark
from pyspark.sql.functions import udf, struct
import stringdist
from pyspark.sql.types import StructType, StructField, StringType, IntegerType, DateType, ArrayType, FloatType, DoubleType, LongType, NullType
from pyspark.ml.linalg import Vectors, VectorUDT
import romantranslateMap = {'A' : 'А', 'B' : 'В', 'C' : 'С', 'E' : 'Е', 'H' : 'Н', 'K' : 'К', 'M' : 'М', 'O' : 'О', 'P' : 'Р', 'T' : 'Т', 'X' : 'Х', 'Y' : 'У'}
column_names = testTable.drop('ID1').drop('ID2').columns
columnsSize = len(column_names)//2def isRoman(numeral):
numeral = numeral.upper()
validRomanNumerals = ["M", "D", "C", "L", "X", "V", "I", "(", ")"]
for letters in numeral:
if letters not in validRomanNumerals:
return False
return Truedef differenceVector(params):
differVector = []
for i in range(0, 3):
if params[i] == None or params[columnsSize + i] == None:
differVector.append(0.0)
elif params[i] == 'НЕТ' or params[columnsSize + i] == 'НЕТ':
differVector.append(0.0)
elif params[i][:params[columnsSize + i].find('-')] == params[columnsSize + i][:params[columnsSize + i].find('-')] or params[i][:params[i].find('-')] == params[columnsSize + i][:params[i].find('-')]:
differVector.append(0.0)
else:
differVector.append(stringdist.levenshtein(params[i], params[columnsSize+i]))
for i in range(3, columnsSize):
# snils
if i == 5 or i == columnsSize + 5:
if params[i] == None or params[columnsSize + i] == None or params[i].find('123-456-789') != -1 or params[i].find('111-111-111') != -1 \
or params[columnsSize + i].find('123-456-789') != -1 or params[columnsSize + i].find('111-111-111') != -1:
differVector.append(0.0)
else:
differVector.append(float(params[i] != params[columnsSize + i]))
# birthCertificate_docNum
elif i == 10 or i == columnsSize + 10:
if params[i] == None or params[columnsSize + i] == None or params[i].find('000000') != -1 or params[i].find('000000') != -1 \
or params[columnsSize + i].find('000000') != -1 or params[columnsSize + i].find('000000') != -1:
differVector.append(0.0)
else:
differVector.append(float(params[i] != params[columnsSize + i]))
# birthCertificate_docSer
elif i == 11 or i == columnsSize + 11:
if params[i] == None or params[columnsSize + i] == None:
differVector.append(0.0)
# check if roman or not, then convert if roman
else:
docSer1 = params[i]
docSer2 = params[columnsSize + i]
if isRoman(params[i][:params[i].index('-')]):
docSer1 = str(roman.fromRoman(params[i][:params[i].index('-')]))
secPart1 = '-'
for elem in params[i][params[i].index('-') + 1:]:
if 65 <= ord(elem) <= 90:
secPart1 += translateMap[elem]
else:
secPart1 = params[i][params[i].index('-'):]
docSer1 += secPart1
if isRoman(params[columnsSize + i][:params[columnsSize + i].index('-')]):
docSer2 = str(roman.fromRoman(params[columnsSize + i][:params[columnsSize + i].index('-')]))
secPart2 = '-'
for elem in params[columnsSize + i][params[columnsSize + i].index('-') + 1:]:
if 65 <= ord(elem) <= 90:
secPart2 += translateMap[elem]
else:
secPart2 = params[columnsSize + i][params[columnsSize + i].index('-'):]
break
docSer2 += secPart2
differVector.append(float(docSer1 != docSer2))
elif params[i] == 0 or params[columnsSize + i] == 0:
differVector.append(0.0)
elif params[i] == None or params[columnsSize + i] == None:
differVector.append(0.0)
else:
differVector.append(float(params[i] != params[columnsSize + i]))
return differVectorfeaturesGenerator = udf(lambda input: Vectors.dense(differenceVector(input)), VectorUDT())
%pyspark
newTestTable = testTable.withColumn('features', featuresGenerator(struct(*column_names))) # all pairs
df = df.withColumn('features', featuresGenerator(struct(*column_names))) # labeled pairs
Split labeled dataframe into training and test dataframes:
%pyspark
from pyspark.ml import Pipeline
from pyspark.ml.classification import RandomForestClassifier
from pyspark.ml.feature import IndexToString, StringIndexer, VectorIndexer
from pyspark.ml.evaluation import MulticlassClassificationEvaluator# split labelled data into two sets
(trainingData, testData) = df.randomSplit([0.7, 0.3])
Train a RF model:
%pyspark
from pyspark.ml.classification import RandomForestClassifierrf = RandomForestClassifier(labelCol='label', featuresCol='features')
pipeline = Pipeline(stages=[rf])
model = pipeline.fit(trainingData)
# Make predictions.
predictions = model.transform(testData)
# predictions.select("predictedLabel", "label", "features").show(5)
Test the RF model:
%pyspark
TP = int(predictions.select("label", "prediction").where((col("label") == 1) & (col('prediction') == 1)).count())
TN = int(predictions.select("label", "prediction").where((col("label") == 0) & (col('prediction') == 0)).count())
FP = int(predictions.select("label", "prediction").where((col("label") == 0) & (col('prediction') == 1)).count())
FN = int(predictions.select("label", "prediction").where((col("label") == 1) & (col('prediction') == 0)).count())
total = int(predictions.select("label").count())print("accuracy = %f" % ((TP + TN) / total))
print("precision = %f" % (TP/ (TP + FP))
print("recall = %f" % (TP / (TP + FN))
How it looks:
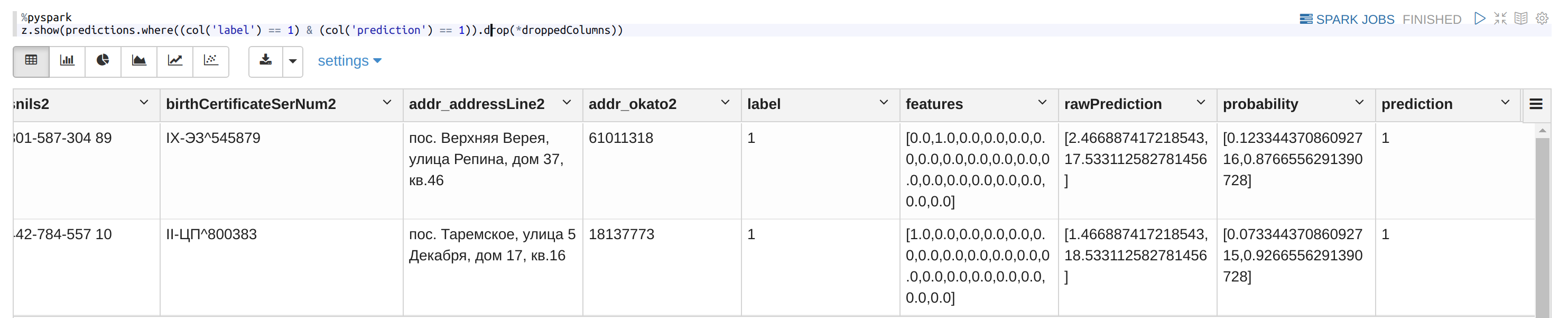
Use the RF model on all the pairs:
%pyspark
allData = model.transform(newTestTable)
Check how many duplicates are found:
%pyspark
allData.where(col('prediction') == 1).count()

Or look at the dataframe:
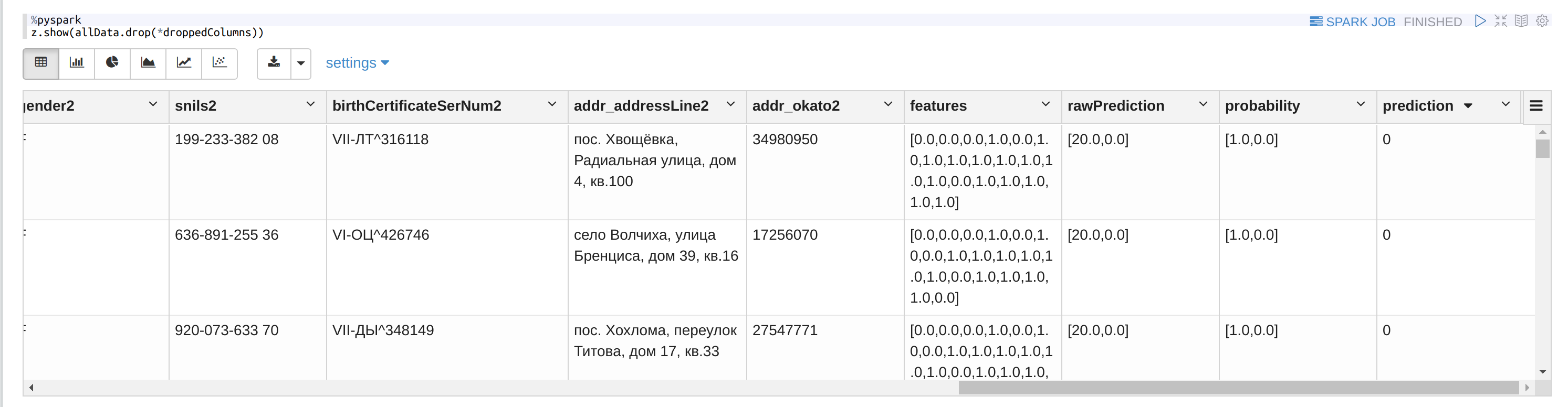
Conclusion
This approach is not ideal. You can make it better by experimenting with feature vectors, a model or increasing the size of labeled dataset.
Also, you can do the same to find duplicates, for example, in shops database, historical research, etc...